Rajendra K. Agrawal, PhD

Structure and Function of Cell’s Protein-Synthesizing “Machine”
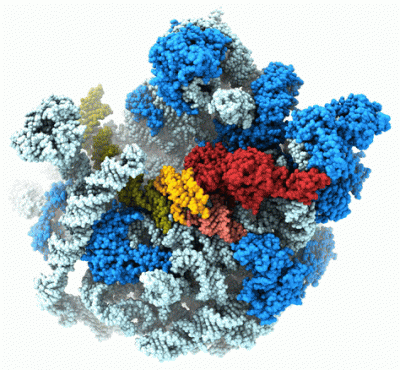
The Agrawal Laboratory studies the mechanism of protein-synthesis, or gene translation, in humans and in pathogenic bacteria. Ribosomes, which lie at the center of the protein synthesis machinery, conduct protein synthesis in all organisms. While the bacterial ribosomes are the major targets of antibiotics, defects in human mitochondrial ribosomes (mitoribosomes) and translation cause multiple devastating genetic diseases. A deep understanding of ribosome structure and function in humans and pathogenic bacteria is necessary for designing or identifying drug targets so that safer drugs can be designed without affecting the human translational machinery. The Agrawal Lab’s emphasis is on human mitochondrial ribosome and ribosomes from pathogenic bacteria. His lab uses biochemical, molecular biology, and high-resolution cryo-electron microscopy[1] (cryo-EM) techniques to study these biological systems.
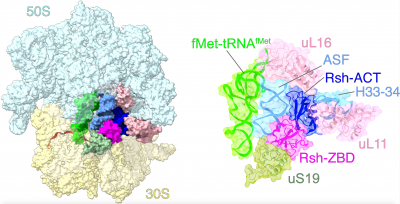
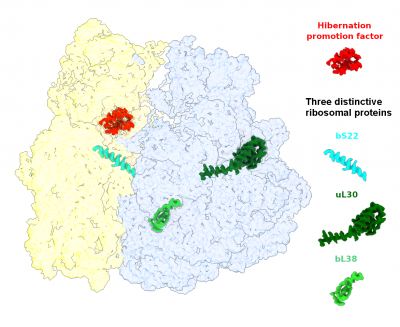
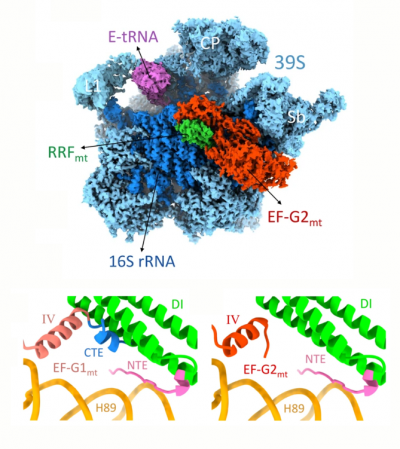
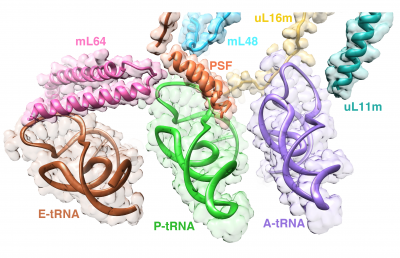
The goal of studies in the Agrawal Lab includes the elucidation of mechanisms by which (i) leaderless mRNAs are recruited to the ribosomes, and (ii) unique component ribosomal proteins and translational factors participate in the process of mitochondrial protein synthesis. His lab was first to determine the structure of a mammalian mitochondrial ribosome.[2] To study mycobacterial protein synthesis, the Agrawal Lab collaborates with other Wadsworth investigators. In collaboration with the Ojha Lab, his lab was first to determine structure of an alternative form of mycobacterial ribosome, revealing unique features of mycobacterial ribosome hibernation[3] that has implications in mycobacterial dormancy and drug resistance. More recent work from his lab, in collaboration with the Ghosh Lab at the Wadsworth Center, revealed a novel molecular mechanism[4] by which the mycobacteria negate the effect of ribosome-binding antibiotics, leading to drug resistance.
Studies in the Agrawal Lab allow him to track conformational transitions undergone by the ribosome and its ligands during protein synthesis. Knowledge of specific conformational transitions is key to understanding the molecular mechanisms of protein synthesis itself, as well as the actions of antibiotics that target the bacterial ribosome or ribosomal ligands to inhibit such transitions. Comparison of results obtained for the bacterial ribosome complexes with those for the host cytosolic and mitochondrial ribosome complexes provides useful information that can lead to identification of new drug targets.