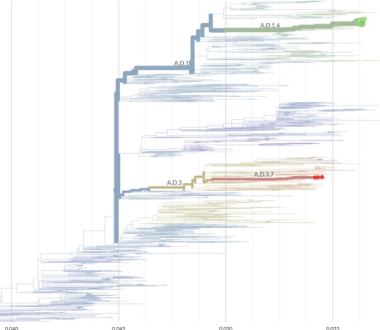
Phylogenetic tree placement of October-November 2025 RSV-A sequences obtained from samples collected in Jefferson County (A.D.1.6, green line) and the Capital Region (A.D.3.7, red line), New York State. Other sequences on the tree, taken from GenBank and Pathoplexus, include global RSV-A reference sequences that represent each of the known RSV-A clades. Image created using NextClade (https://clades.nextstrain.org). Data, write up and figure supplied by Sara Griesemer and Daryl Lamson, Research and Development Group, Laboratory of Viral Diseases.
The New York State Department of Health’s Wadsworth Center and Division of Epidemiology are collaborating on the investigation of a Respiratory Syncytial Virus (RSV) outbreak in northern New York State. RSV primarily causes clinically significant illness in children under five years of age and older adults, often presenting as influenza-like illness or pneumonia, and commonly requiring hospitalization in severe cases.
The Wadsworth Center’s Laboratory of Viral Diseases conducted initial testing to distinguish between the two RSV subtypes (RSV-A and RSV-B) and performed whole-genome sequencing (WGS) on RSV-positive samples from Jefferson and Lewis counties, where a notable increase in cases was identified. Subtyping revealed predominant circulation RSV-A in these northern counties, and sequencing confirmed the presence of a single RSV-A lineage, A.D.1.6. This lineage has been less commonly reported in the United States, accounting for fewer than 20 percent of RSV cases submitted to national databases over the past two years. In contrast, specimens from other counties in the Capital Region have shown predominant circulation of the RSV-B subtype. The limited RSV-A detections in this region belong to a different lineage, A.D.3.7.
Statewide investigation of circulating RSV strains is ongoing to provide epidemiologic insight to support public health response and surveillance efforts.