Welcome to the Interactive Genomics home page of the Wadsworth Center. This page contains links to genomic views of bacteria in which we display RNA profiling and ChIP-seq data mapped to reference genomes. RNA profiles include RNA-seq, transcription start site (TSS) and Ribo-seq data. Combining these data will allow the user to accurately determine gene boundaries, identify novel genes, leadered and leaderless transcripts, and non-coding RNAs. The Salmonella page includes ChIP-seq data for mapping transcription-factor binding sites, and corresponding RNA-seq data. The data are displayed using JBrowse, developed by Skinner et al., JBrowse: A next generation genome browser. Genome Research. 2009;19:1630-1638.
Interactive Genomes
To determine the gene architecture of your favourite gene:
- Select your organism.
- At the left of each page is a list of buttons that allow selection of the relevant data. Check the reference sequence you wish to use (e.g., mc2155) and the genome features you wish. These will automatically populate the browser in order of selection.
- Select the appropriate RNA data sets for each strand. These will vary for each organism, and may include RNA-seq, Ribo-seq, TSS and ChIP-seq data. For each organism, there are biological replicates (Rep1, Rep2). These show very little variation, so we recommend selecting only one data set. For some genomes (e.g., Salmonella), there are also multiple conditions or genetic backgrounds from which RNA was collected and analyzed. Chose your favourite conditions(s). Note the page loads as you click so we recommend selecting the plus-strand information first, and then follow the same format for the minus-strand information. In the example below, we have selected RNA-seq and then Ribo-seq data from the positive-strand first , so it is easier to align reads with each other and the annotated genome.
- To browse the genome you can use the arrows in the header. Alternatively, enter a nucleotide coordinate or a gene number into the box and select “Go”. You can also click on a gene, hold the click, and drag left or right.
- Zoom in and out using the -/+ buttons, which will take you down to the nucleotide level.
- The Y-axis scale is arbitrarily set at 500 mapped reads at any given map position of the genome. If the read depth is off scale, that area of the peak will change colour (e.g., from blue to red for sense strand). Slide the mouse over the peak and it will tell you the number of reads. The scale can be adjusted as follows: click on the track title and then the downward-pointing triangle (on the right). Select “edit config”. Change "max_score": 500, to whatever number you want, and then click “apply”.
Acknowledgments
Please cite this page if you use this information for publications. We acknowledge support from: the Wadsworth Center; NIAID grants AI09719103, AI117158; NIGMS grant 1DP2OD007188, and NIAID contract HHSN272200900040C.
Associated Publications
smegmatis.wadsworth.org and mtb.wadsworth.org
Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA (2015) Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape. PLoS Genet 11(11): e1005641
salmonella.wadsworth.org
Smith, C., Stringer, A. M., Mao, C., Palumbo, M. J. and Wade, J. T. Mapping the Regulatory Network for Salmonella Typhimurium Invasion. Submitted to PLoS Pathog.
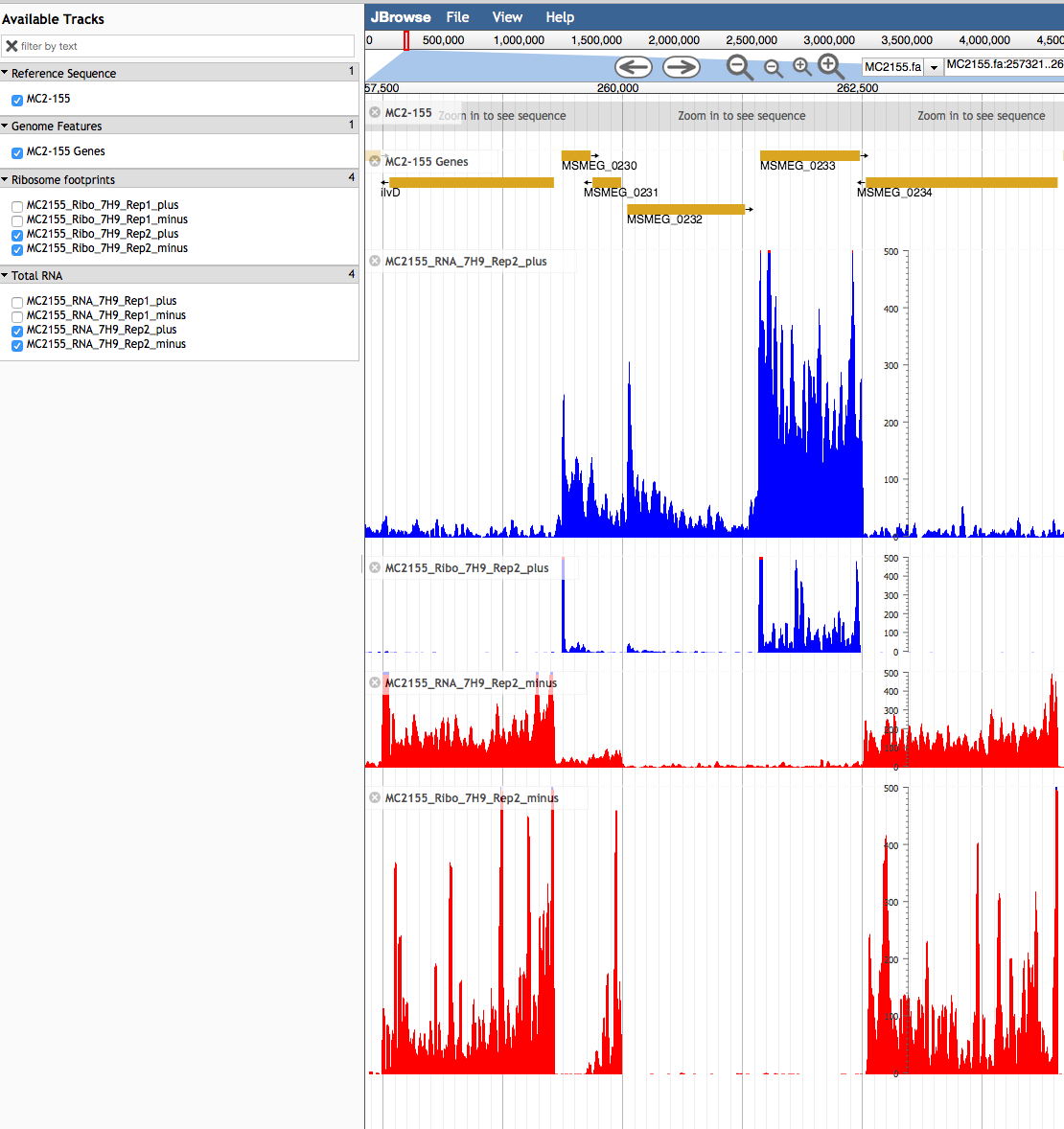
The first two plots, in blue, show the transcriptional (RNA-seq) and translational (Ribo-seq) profiles for genes expressed left to right (0230, 0232 and 0233). The two bottom panels, in red, show the profiles for genes expressed from right to left (0234, 0231 and 0229). By examining the data it is possible to map the likely transcriptional and translational start site for each gene.