Research Interests
Our research explores the molecular biology of bacterial viruses (phages) and other mobile genetic elements, with a focus on how they shape the ecology and evolution of bacterial pathogens. Just as our lives are shaped by viral epidemics, bacteria face a constant battle against viruses, shaping their genetic diversity and influencing their ability to cause disease and resist antibiotics. Studying how phages interact with their bacterial hosts provides fundamental insights into the biology of those bacteria. After all, phages (and other genetic parasites) have been studying bacteria for billions of years longer than we have! By understanding these phage-bacteria interactions, we can better predict evolutionary outcomes, and ultimately derive strategic interventions to guide the evolution of bacteria towards favorable outcomes.
Currently, our lab is pioneering novel strategies to isolate phages with exciting and potentially exploitable characteristics, such as dependency on antibiotic resistance plasmids. We are studying the biology of these unusual plasmid-dependent phages, as well as the plasmids they depend on, gaining unique insights into the molecular biology of antibiotic resistant bacterial pathogens.
Plasmid-dependent phages
Across the world, bacterial infections are becoming increasingly difficult to treat due to the growing prevalence of antibiotic resistance. The primary mechanism responsible for the emergence of antibiotic resistance in pathogenic bacteria is horizontal gene transfer, wherein genes encoding antibiotic resistance are transferred between distantly-related bacteria. Conjugative plasmids are the key genetic elements driving horizontal gene transfer because they are autonomously replicative and mobile, encoding genes necessary for their own maintenance and transfer. Conjugative plasmids generate a large secretory pilus, a nanomachine on the surface of the bacterial cell that permits horizontal transfer of plasmid DNA into new hosts. Thus, antibiotic resistance can travel from harmless bacteria into deadly pathogens.
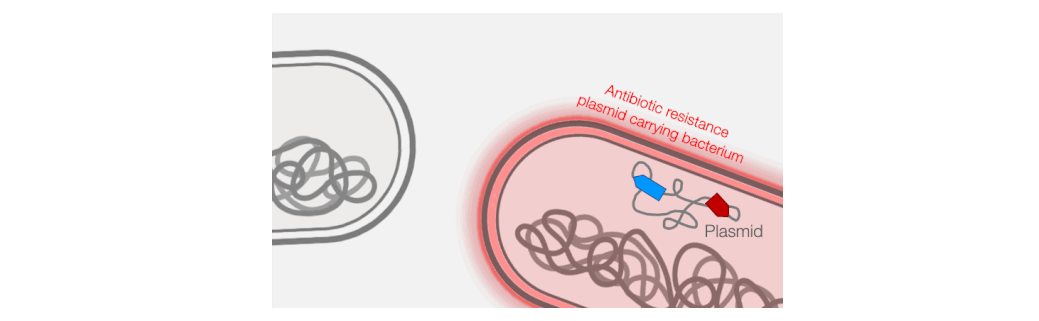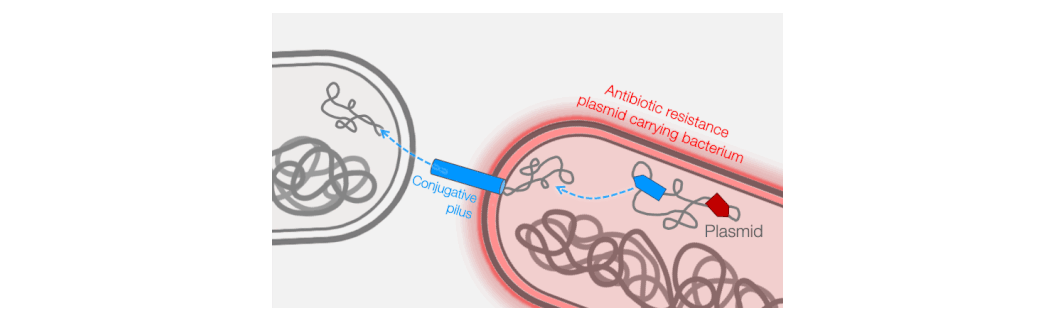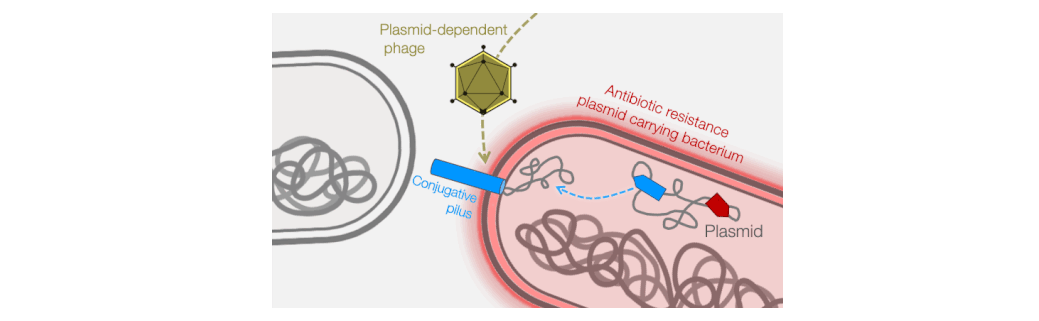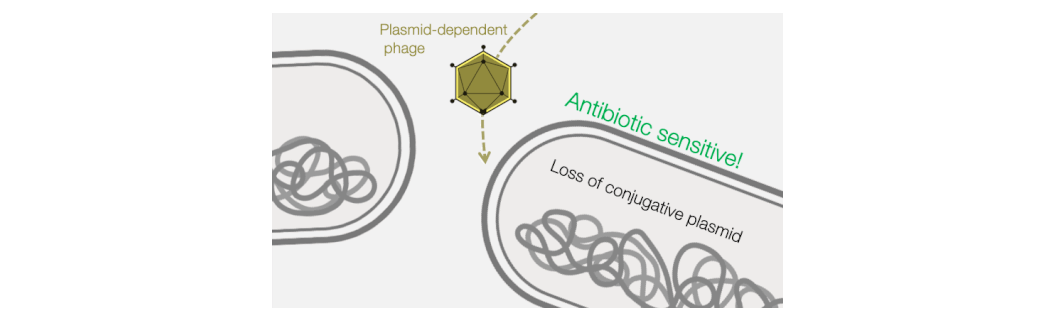
Fortunately, phages provide an excellent strategy for limiting pilus-mediated spread of antibiotic-resistance plasmids. Phages identify their hosts by interacting with receptors on the cell surface, and the conjugative pilus provides a landing pad for phages. Therefore, the very system that plasmids use to mediate horizontal gene transfer represents an “Achilles heel” of diverse antibiotic resistant bacterial pathogens. We seek to exploit that weakness by identifying and characterizing plasmid-dependent phages (PDPs) that infect and destroy bacteria with conjugative plasmids. Research using the few known PDPs shows they can effectively reduce the occurrence and transmission of antibiotic resistance in bacterial populations. This is because PDPs impose an evolutionary trade-off between plasmid-mediated resistance to antibiotics and phage sensitivity. To understand the real-life significance and potential applications of this phage vs antibiotic-resistance trade-off, we are studying how widespread and abundant PDPs are in nature, their fundamental molecular biology, and the evolutionary impact of phage-plasmid interactions on bacteria.
Join us!
We admit PhD or Masters students through the BMS program at UAlbany. Currently enrolled students should contact Siân about rotating in the lab. For postdoctoral or other roles, contact Siân by email to discuss current opportunities.
